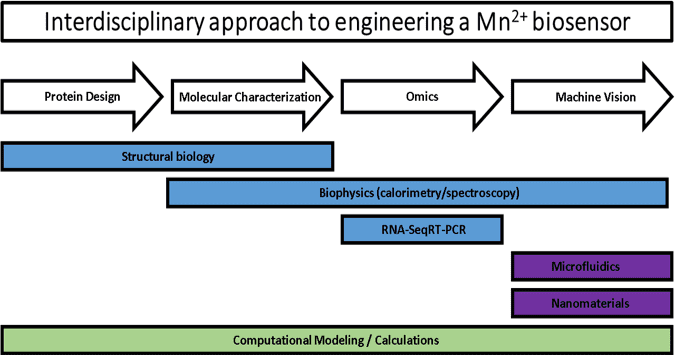
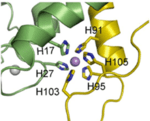
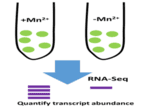
Acknowledgement: The following Publications and Master’s Theses were made possible through support from the US National Science Foundation through the CREST program’s Award Number: HRD 1547757, Fisk University Center of Excellence for Biological Signatures and Sensing (BioSS)
Publications
- Eric Lukosi, Elan Herrera, Daniel Hamm, Kyung-Min Lee, Brenden Wiggins, Pavel Trtik, Dayakar Penumadu, Stephen Young, Louis Santodonato, Hassina Bilheux, Arnold Burger, Liviu Matei, Ashley C Stowe. "Lithium indium diselenide: A new scintillator for neutron imaging," Nuclear Instruments and Methods in Physics Research Section A, v.830, 2016, p. 104-149. https://doi.org/10.1016/j.nima.2016.05.063
- G Calvert, C Guguschev, A Burger, M Groza, JJ Derby, RS Feigelson. "High speed growth of SrI2 scintillator crystals by the EFG process," Journal of Crystal Growth, v.455, 2016, p. 143-151. https://doi.org/10.1016/j.jcrysgro.2016.10.024
- David Caudel, Michael McCurdy, Daniel M Fleetwood, Robert A Reed, Robert A Weller, Brandon Goodwin, Emmanuel Rowe, Vladimir Buliga, Michael Groza, Keivan Stassun, Arnold Burger. "Radiation damage of strontium iodide crystals due to irradiation by 137 Cs gamma rays: A novel approach to altering nonproportionality," Nuclear Instruments and Methods in Physics Research Section A: Accelerators, Spectrometers, Detectors and Associated Equipment, v.835, 2016, p. 177-181. https://doi.org/10.1016/j.nima.2016.08.041
- Elan Herrera, Daniel Hamm, Brenden Wiggins, Rob Milburn, Arnold Burger, Hassina Bilheux, Louis Santodonato, Ondrej Chvala, Ashley Stowe, Eric Lukosi. "Arnold Burger Arnold Burger LISe pixel detector for neutron imaging," Nuclear Instruments and Methods in Physics Research Section A: Accelerators, Spectrometers, Detectors and Associated Equipment, v.833, 2016, p. 142-148. https://doi.org/10.1016/j.nima.2016.07.035
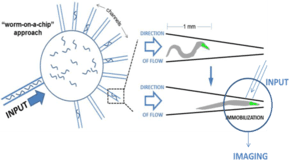
- Lijie Yang, Tao Hong, Yin Zhang, Jose G Sanchez Arriola, Brian L. Nelms, Richard Mu, Deyu Li. "A highly efficient microfluidic diode for sorting and immobilization of C. elegans," Biomedical Microdevices, 2017. https://doi.org/10.1007/s10544-017-0175-2
- Emmanuel Jackson, Saffron Little, Dana Franklin, Jennifer A. Gaddy, Steven M. Damo. "Expression, Purification and Antimicrobial Activity of S100A12," Journal of Visualized Experiments, 2017. https://doi.org/10.3791/55557
- A. Lakhtakia, D. E. Wolfe, M. W. Horn, J. Mazurowski, A. Burger, P. P. Banerjee. "Bioinspired multicontrollable metasurfaces and metamaterials for terahertz applications," SPIE Proceedings, v.10162, 2017, p. 101620V. https://doi.org/10.1117/12.2258683
- Pokhrel, A. Burger, M. Groza, Y. Mao. "Enhance the photoluminescence and radioluminescence of La 2 Zr 2 O 7: Eu 3+ core nanoparticles by coating with a thin Y 2 O 3 shell," Optical Materials, v.68, 2017, p. 35. https://doi.org/10.1016/j.optmat.2016.11.00 8
- N. J. Cherepy, P. R. Beck, S. A. Payne, E. L. Swanberg, B. M. Wihl, S. E. Fisher, S. Hunter, P. A. Thelin, C. J. Delzer, S. Shahbazi, A. Burger, K. S. Shah, R. Hawrami, L. A. Boatner, M. Momayezi, K. Stevens, M. H. Randles, D. Solodovnikov. "History and current status of strontium iodide scintillators," SPIE Proceedings, v.10392, 2017, p. 1039202.
https://doi.org/10.1117/12.2276302
- A. C. Stowe, J. Preston, J. Bell, A. Burger. "Lithium indium diselenide pressed ceramics," SPIE Proceedings, v.10392, 2017, p. 103920N. https://doi.org/10.1117/12.2276275
- B. Wiggins, J. Bell, J. Woodward, B. Goodwin, K. Stassun, A. Burger, A. Stowe,. "Crystal growth of LiIn1? xGaxSe2 crystals," Journal of Crystal Growth, v.468, 2017, p. 326. https://doi.org/10.1016/j.jcrysgro.2016.10.051
- J. G. Bodnarik, D. K. Hamara, A. Burger, V. Buliga, J. C. Egner, M. Groza, W M. Harris, L. Matei, K. G. Stassun, A. C. Stowe. "Neutron detector development for microsatellites," SPIE Proceedings, v.10392, 2017, p. 103920M. https://doi.org/10.1117/12.2275682
13.G. Cooper, A. Stowe, J. Preston, B. Wiggins, K. Stassun, A. Burger. "The Characterization and Optimization of Lithium Hafnium Chloride Crystal Scintillators for Neutron Detection," SciTech Connect, v.IROS172, 2017. https://doi.org/10.2172/1410677
- C. Cardenas, A. Burger, B. Goodwin, M. Groza, M. Laubenstein, S. Nagorny, E. Rowe. "Pulse-shape discrimination with Cs2HfCl6 crystal scintillator," Nuclear Instruments and Methods in Physics Research, v.869, 2017, p. 63. https://doi.org/10.1016/j.nima.2017.06.041
- S. Sahi, M. Groza, W. Zhang, P. A. Do, R. Kenarangui, A. Burger, J.Zhang, W. Chen. "High-loaded and transparent La x Ce1-x F3?polystyrene nanocomposite scintillators for radiation detection," Canadian Journal of Chemistry, v.95, 2017, p. 1233. https://doi.org/10.1139/cjc-2017-0211
- C. Cardenas, A. Burger, M. L. DiVacri, B. Goodwin, M. Groza, M. Laubenstein, S. Nagorny, S. Nisi, E. Rowe. "Internal contamination of the Cs2HfCl6 crystal scintillator," Nuclear Instruments and Methods in Physics Research Section A, v.872, 2017, p. 23. https://doi.org/10.1016/j.nima.2017.08.006
- E. D. Lukosi, E. H. Herrera, D. S. Hamm, A. Burger, A. C. Stowe. "Neutron imaging with lithium indium diselenide: Surface properties, spatial resolution, and computed tomography," Nuclear Instruments and Methods in Physics Research, v.872, 2017, p. 181. https://doi.org/10.1016/j.nima.2017.08.028
- E. Herrera, D. Hamm, A. Stowe, J. Preston, B. Wiggins, A. Burger, E. Lukos. "Neutron Imaging with Timepix Coupled Lithium Indium Diselenide," Journal of Imaging, v.4, 2017, p. 10. https://doi.org/10.3390/jimaging4010010
- S. Lam, C. Guguschev, A. Burger, M. Hackett, S. Motakef. "Crystal growth and scintillation performance of Cs2HfCl6 and Cs2HfCl4Br2," Journal of Crystal Growth, v.483, 2018, p. 12, https://doi.org/10.1016/j.jcrysgro.2017.11.013
- E. Brown, Z. D. Fleischman, L. D. Merkle, E. Rowe, A. Burger, S. A. Payne, M. Dubinskii. "Optical spectroscopy of holmium doped K2LaCl5," Journal of Luminescence, v.196, 2018, p. https://doi.org/10.1016/j.jlumin.2017.12.040
- Sharee N. Brewer,Tykeena Watson, Qingxia Li, Xinyao Yang. "Optimal Scheduling for CK Advertisements on TV Programs," Proceedings of the National Conference On Undergraduate Research (NCUR), 2017, p. 1235.
22.D. S. Hamm, M. Rust, E. H. Herrera, L. Matei, V. Buliga, M. Groza, A. Burger, A. Stowe, J. Preston, E. D. Lukosi. "Semiconducting lithium indium diselenide: Charge-carrier properties and the impacts of high flux thermal neutron irradiation," Applied Physics Letters, v.112, 2018, p. 242104. https://doi.org/10.1063/1.5028269
- E. Brown, Z. D. Fleischman, L. D. Merkle, E. Rowe, A. Burger, S. A. Payne, M. Dubinskii,. "Optical spectroscopy of holmium doped K2LaCl5," Journal of Luminescence, v.196, 2018, p. 221. https://doi.org/10.1016/j.jlumin.2017.12.040
- E. Brown, Z. Fleischman, L. Merkle, E. Rowe, A. Burger, S. Payne, M. Dubinskiy. "Infrared absorption and fluorescence properties of Holmium doped Potassium Lanthanum Chloride," International Society for Optics and Photonics, v.10637, 2018, p. 10670X. https://doi.org/10.1117/12.2309578
- E. Lukosi, D. Onken, D. Hamm, C. Brown, A. V. Ievlev, A. Burger, J. Preston, R. Williams, A. Stowe. "Intrinsic lithium indium diselenide: Scintillation properties and defect states," Journal of Luminescence, v.205, 2019, p. 346. https://doi.org/10.1016/j.jlumin.2018.09.023
- Ghosh, S., Garcia, V., Singewald, K., Damo, S.M., Saxena, S.. "Cu (II) EPR Reveals Two Distinct Binding Sites and Oligomerization of Innate Immune Protein Calgranulin C," Applied Magnetic Resonance, v.49, 2018, p. 1299. https://doi.org10.1007/s00723-018-1053-7
- Janette M. Shank, Brittni R. Kelley, Joseph W. Jackson, Jessica L. Tweedie, Dana Franklin,Steven M. Damo, Jennifer A. Gaddy, Caitlin N. Murphy, and Jeremiah G. Johnson. "The host antimicrobial protein calgranulin C 1 participates in the control of Campylobacter 2 jejuni growth via zinc sequestration," Infection and Immunity, v.86, 2018, p. 1. https://DOI.org/10.1128/IAI.00234-18
- Kozlyuk, N., Monteith, A.J., Garcia, V., Damo, S.M., Skaar, E.P., Chazin, W.J.. "S100 Proteins in the Innate Immune Response to Pathogens," Methods in Molecular Biology, v.1929, 2019, p. 275. https://doi.org/10.1007/978-1-4939-9030-6_18
- Li, Q. & Yang, X. "Integrated Case Studies in Teaching Introductory Mathematics Courses at College," European Journal of Educational Sciences, v.5, 2018, p. 1. https://doi.org/10.19044/ejes.v5no3a1
- Li, Q., Yang, X. "On Suprememum of a set in a Dedikind Complete Topological Space," JOURNAL OF ADVANCES IN MATHEMATICS, v.14, 2018, p. 8025. https://doi.org/10.24297/jam.v14i2.7807
- Shank, J.M., Kelley, B.R., Jackson, J.W., Tweedie, J.L., Franklin, D., Damo, S.M., Gaddy, J.A., Murphy, C.N., Johnson, J.G.. "The host antimicrobial protein calgranulin C participates in the control of Campylobacter jejuni growth via zinc sequestration.," Infection and immunity, v.86, 2018. https://doi.org/10.1128/IAI.00234-18
- Y. He, L. Matei, H. J. Jung, K. M. McCall, M. Chen, C. C. Stoumpos, Z. Liu, J. A. Peters, D. Y. Chung, B. W. Wessels, M. R. Wasielewski, V. P. Dravid, A. Burger, M. G. Kanatzidis. "High spectral resolution of gamma-rays at room temperature by perovskite CsPbBr3 single crystals," Nature Communications, v.9, 2018, p. 1609. https://doi.org/10.1038/s41467-018-04073-3
- E. Rowe, W. B. Goodwin, P. Bhattacharya, G. Cooper, N. Schley, M. Groza, N. J. Cherepy, S. A. Payne, A. Burger. "Preparation, Structure and Scintillation of Cesiu m Hafnium Chloride Bromide Crystals," Journal of Crystal Growth, v.509, 2019, p. 124-128 https://doi.org/10.1016/j.jcrysgro.2018.08.033
- Elsa Ariesanti, Rastgo Hawrami, Arnold Burger, Shariar Motakef. "Improved growth and scintillation properties of intrinsic, non-hygroscopic scintillator Cs2HfCl6," Journal of Luminescence, v.217, 2020, p. 116784. https://doi.org/10.1016/j.jlumin.2019.116784
- Ralph B James, Arnold Burger, Stephen A Payne. "Hard X-Ray, Gamma-Ray, and Neutron Detector Physics XXI," Proc. of SPIE Vol, v.11114, 2019, p. 1111401. http://toc.proceedings.com/51710webtoc.pdf
- Rastgo Hawrami, Elsa Ariesanti, Vlad Buliga, Arnold Burger. "Thallium strontium iodide: A high efficiency scintillator for gamma-ray detection," Optical Materials, v.100, 2020, p. 109624. https://doi.org/10.1016/j.optmat.2019.109624
- Rastgo Hawrami, Elsa Ariesanti, Vlad Buliga, Liviu Matei, Shariar Motakef, Arnold Burger. "Advanced high-performance large diameter Cs2HfCl6 (CHC) and mixed halides scintillator," Journal of Crystal Growth, v.533, 2020, p. 125473. https://doi.org/10.1016/j.jcrysgro.2019.125473
- Rastgo Hawrami, Elsa Ariesanti, Vladimir Buliga, Arnold Burger, Stephanie Lam, Shariar Motakef. "Tl2HfCl6 and Tl2ZrCl6: Intrinsic Tl-, Hf-, and Zr-based scintillators," Journal of Crystal Growth, v.531, 2020, p. 125316. https://doi.org/10.1016/j.jcrysgro.2019.125316
- Steven Damo, Jennifer Gaddy. "S100A12 in Digestive Diseases and Health: A Scoping Review," Gastroenterology Research and Practice, v.2020, 2020, p. 868373. https://doi.org/10.1155/2020/2868373
- Steven Damo, Rekha Pattanayek, Chris Johnson. "The CaMKII inhibitor KN93-calmodulin interaction and implications for calmodulin tuning of NaV1. 5 and RyR2 function," Cell Calcium, v.82, 2019, p. 102063. https://doi.org/10.1016/j.ceca.2019.102063
- Steven Damo, Walter Chazin, Eric Skaar. "S100 Proteins in the Innate Immune Response to Pathogens," Methods in Molecular Biology, v.1929, 2019, p. 275. https://doi.org/10.1007/978-1-4939-9030-6_18
Master’s Theses
1) Jodiene Johnson, “Deciphering the role of Germline-Specific Protein MAGEA9 in Cancer”, Master’s thesis, Fisk University, John Hope and Aurelia E. Franklin Library, 2020.
2) Murtidevi Nauth, “THE EFFECT OF BINARY BLACK HOLES ON GRAVITATIONAL WAVE OBSERVATIONS OF THE VERY EARLY UNIVERSE”, Master’s thesis, Fisk University, John Hope and Aurelia E. Franklin Library, 2020.
3) Bria Collier, “CHARACTERIZING THE MOISTURE SENSITIVITY OF Cs2HfCl6 AND Cs2HfCl4Br2”, Master’s thesis, Fisk University, John Hope and Aurelia E. Franklin Library, 2020.
4) Krystal Ruiz-Rocha, “A Sea of Black Holes: Characterizing the Expected Signature of Stellar Origin Binaries in the LISA Band”, Master’s thesis, Fisk University, John Hope and Aurelia E. Franklin Library, 2020.
5) RaNashia Boone, “Understanding antibiotic resistance mechanisms in Acinetobacter baumannii”, Master’s thesis, Fisk University, John Hope and Aurelia E. Franklin Library, 2020.
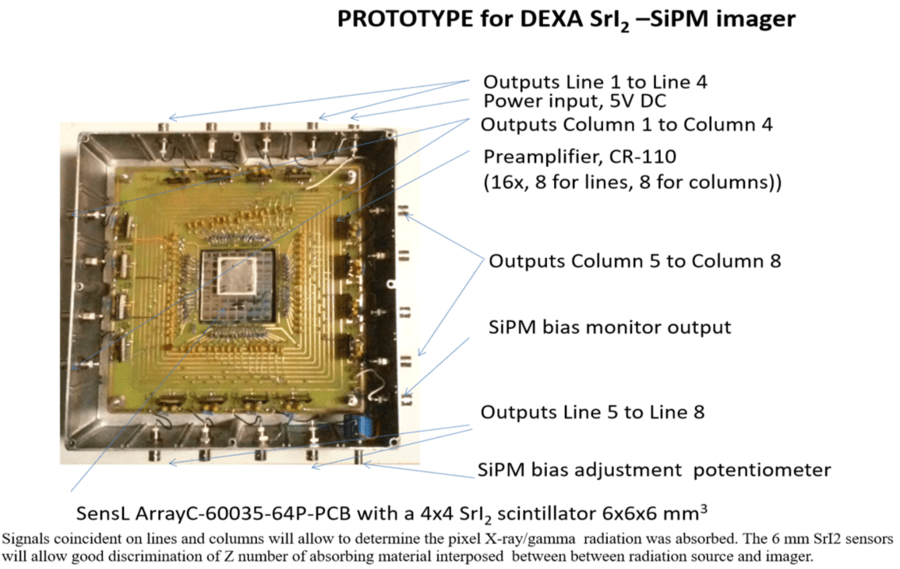
6) Michelle Gomez, “Calibration of a DEXA Prototype Using Strontium Iodide Doped with Europium Coupled to a Silicon Photomultiplier for Bone Imaging”, Master’s thesis, Fisk University, John Hope and Aurelia E. Franklin Library, 2020.
7) Kricia Ruano Espinoza, “Europium-doped strontium iodide (SrI2(Eu2+)) scintillator crystals in dual energy x-ray absorptiometry (DEXA) for bone densitometry imaging”, Master’s thesis, Fisk University, John Hope and Aurelia E. Franklin Library, 2020.
8) Aaron Hunsaker, “Production and Characterization of Ceramic Cs2HfCI6 for Gamma Ray Spectroscopy”, Master’s thesis, Fisk University, John Hope and Aurelia E. Franklin Library, 2019.
9) Matthew Galazzo, “Post Synthesis Modifiable Polymers sing Cyanuric Chloride and Derivatives for Fuel Cell Applications”, Master’s thesis, Fisk University, John Hope and Aurelia E. Franklin Library, 2019
10) Crisel Suarez, “Computational Methods for the Scintillator Cs2HfCl6 (CHC) and Solar Flares”, Master’s thesis, Fisk University, John Hope and Aurelia E. Franklin Library, 2019.
11) Briana Whitehead, “Elucidating the Molecular Mechanisms of Antibiotic Resistance in the Human Pathogen Acinetobacter baumannii”, Master’s thesis, Fisk University, John Hope and Aurelia E. Franklin Library, 2019.
12) Amber Britt, “Photochemical Modeling of Terrestrial Atmospheres and Their Potential Biosignatures: The Search for Extraterrestrial Life”, Master’s thesis, Fisk University, John Hope and Aurelia E. Franklin Library, 2019.
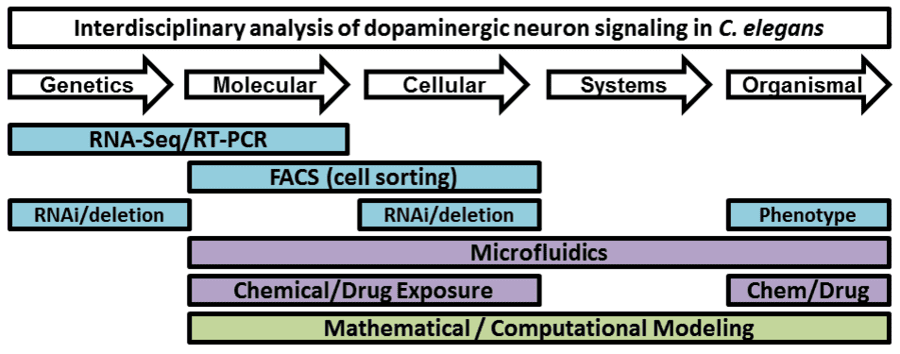
13) Austin Anthony, “RNA sequencing- a multipurpose tool for identifying genes critical for dopamine system functioning or bacterial resistance to radiation”, Master’s thesis, Fisk University, John Hope and Aurelia E. Franklin Library, 2019.
14) Shawndise Beachem, “Study of the potential of europium doped strontium iodide in comparison to sodium iodide for optimum gamma camera parameters for SPECT imaging”, Master’s thesis, Fisk University, John Hope and Aurelia E. Franklin Library, 2018.
15) Debresha Shelton, “Monoamine oxidase and catechol-O-methyltranserases Modulate Dopamine-Mediated Behavior”, Master’s thesis, Fisk University, John Hope and Aurelia E. Franklin Library, 2018.
16) Destane Garrett, “FKH-8 Controls Transcription of Important Signaling Components for CO2-Sensing by BAG Neurons”, Master’s thesis, Fisk University, John Hope and Aurelia E. Franklin Library, 2018.


 Fisk faculty participating in the Center research and professional skills development activities and a number of additional faculty members at Vanderbilt and national laboratories collaborators who are also engaged in the training of graduate students participating in the Fisk-Vanderbilt Master’s-to-Ph.D. Bridge program.
Fisk faculty participating in the Center research and professional skills development activities and a number of additional faculty members at Vanderbilt and national laboratories collaborators who are also engaged in the training of graduate students participating in the Fisk-Vanderbilt Master’s-to-Ph.D. Bridge program.